Breaking down the microbiology world one bite at a time
Technology to tackle viral infections
Oxford Nanopore sequencing is a technology that can be used to read the genetic material (DNA or RNA) of viruses. This technology can help in monitoring viral infections because it can quickly and accurately identify the specific type of virus causing the infection.
Traditionally, viral infections are monitored by taking a sample from a patient and then using various methods to identify the virus (such as a PCR). However, these methods can be time-consuming and may not always provide accurate results. Oxford Nanopore sequencing, on the other hand, can provide fast and accurate results, making it a useful tool for monitoring viral infections.
This method is fast and accurate because it is a real-time sequencing technology that directly reads the DNA or RNA molecule as it passes through a tiny pore or channel.
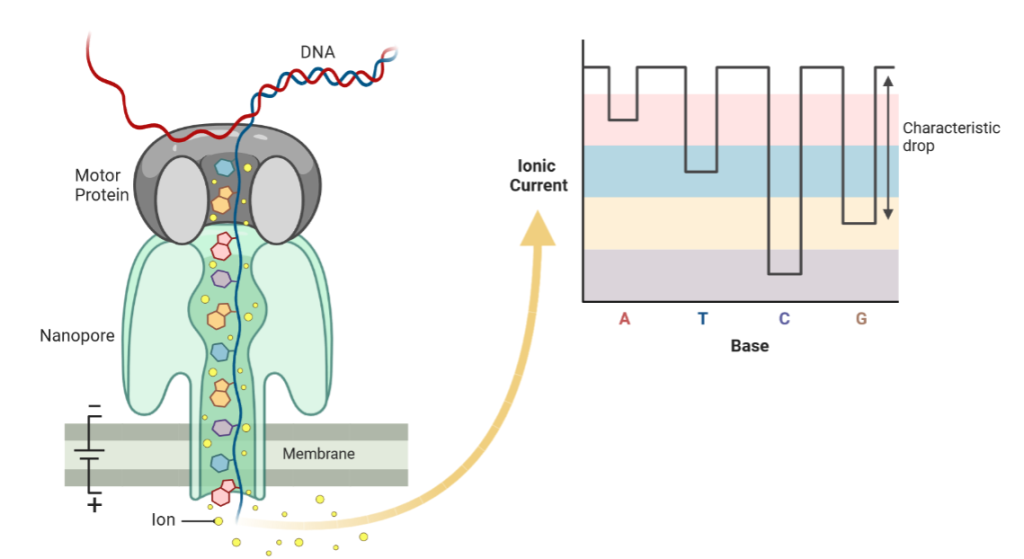
The DNA or RNA molecule is electrically charged, and as it passes through the pore, it causes changes in the electrical current, which can be detected and analysed in real time. Additionally, because Oxford Nanopore sequencing directly reads the DNA or RNA molecule, it can provide longer read lengths and a higher degree of accuracy compared to other sequencing technologies. This is because other sequencing methods may require amplification or fragmentation of the DNA or RNA, which can introduce errors or biases in the resulting sequence data.
But how can we use it to monitor viral infections?
In their study, Munroe and colleagues developed a method to more efficiently and cost-effectively sequence the genetic material of viruses, such as the SARS-CoV-2 virus responsible for the COVID-19 pandemic. Many different labs have looked at using nanopore sequencing to target viral infections even in animals to provide faster and more accurate diagnoses to farmers.
One challenge of using this sequencing technology is obtaining enough DNA or RNA to run the machine accurately. In this study, the authors developed an experimental procedure to predict when sufficient DNA or RNA data were obtained from each sample for high-quality genetic data. To further improve the efficiency of Oxford Nanopore sequencing, the researchers developed a tool that automates adaptive sampling on the sequencing machine during the sequencing process. Adaptive sampling is a way of selecting and sequencing specific molecules for analysis. Instead of using traditional methods to target a DNA or RNA sequence of interest, such as depletion or enrichment of the sequence in a lab, Nanopore has developed a software-controlled approach. This allows researchers to control individual pores and request the ejection of a specific molecule being sequenced in a pore. In other words, users can stop collecting the sequence data from a DNA or RNA molecule on demand. This helps to balance the amount of sequencing coverage across different samples and regions of the virus’s genetic material, reducing the time needed to obtain complete genetic sequences of the virus.
In other words, adaptive sampling allows testing for the SARS-CoV-2 virus in a faster and more efficient way because the SARS-CoV-2 sequences can be targeted directly. By using this method the team was able to get the sequences of the viral genomes in a shorter time, and only run the samples until necessary, allowing a faster turnover. While saving time, the authors showed that it doesn’t affect the accuracy of the results. For now, 96 samples can be tested using this method, and we may be able to test even more in the future!
Overall, these improvements in sequencing efficiency and accuracy are important for monitoring and surveillance of viral pathogens, as well as for developing treatments and vaccines to combat infectious diseases like COVID-19.
Featured image: made with biorender template