Breaking down the microbiology world one bite at a time
Cancer-causing bacteria
A report from the National Cancer Institute in 2020 estimated that 1.8 million cases of cancer with 600,000 deaths would occur in the United States every year. When scientists discuss carcinogens, or cancer-causing agents, a few common ones come to mind. Some widely known carcinogens include smoking tobacco, alcohol, and ultraviolet light from the sun.
However, many people don’t often think of bacteria as potential carcinogens. This is mostly because bacteria often cause acute, short infections that are cleared by antibiotics and the immune system fairly quickly. However, some bacterial infections can have long lasting effects on the immune system of the host, such as the development of cancer.
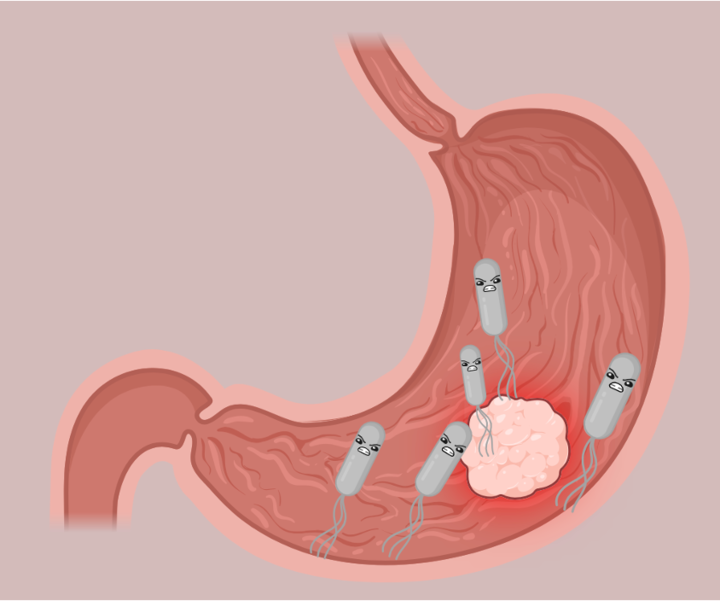
The most common bacterium associated with cancer is the stomach pathogen Helicobacter pylori, which has been classified as a carcinogen since 1994. Despite the correlation between H. pylori and gastric cancer, it is not fully understood how this relationship works. Some studies suggest that H. pylori promotes cancer through prolonged inflammation, which in turn increases the likelihood that a person will develop cancer. However, the authors of this paper wondered if specific features of the H. pylori strains themselves could promote cancer more than other strains.
Looking for mutations at the source.
To identify mutations in H. pylori strains potentially promoting gastric cancer, the researchers in this study utilized a vast collection of sequenced clinical isolates of H. pylori from around the world.
When looking at the DNA sequences of these strains, the authors found that one particular mutation in a bacterial protease called high temperature requirement A (HtrA) was extremely common in H. pylori strains isolated from patients with severe forms of gastric cancer.
Interestingly this mutation was a single nucleotide polymorphism (SNP), which is when a single nucleotide is changed in a gene. Usually, a SNP doesn’t affect the overall protein sequence of a gene that often. This is because the codons that encode each amino acid often have multiple variations.
However, the single nucleotide change identified here was significant, as it resulted in an amino acid change in the 171st amino acid of the protein. Since in general, protein sequence is important for its structure and function, the authors were curious if this part of the protein sequence was important for its function.
Indeed, the amino acid was in a region of HtrA responsible for its activity–the protease domain. Proteases are often identified as virulence factors of pathogens, since their function is to break down proteins, including ones that are important for the host. This is especially the case with HtrA, which is known to break down tight junctions in stomach cells (Figure 1). This cleavage allows H. pylori to squeeze in between the stomach cells to an area far away from stomach acid, which is too acidic for bacteria to grow. Once in a favorable environment, H. pylori can focus its energy into injecting toxins into the stomach cells, which activate the immune response and eventually promote cancer development.
What does this mutation do?
Now that this mutation was identified, the authors wondered if this mutation actually affected the activity of HtrA, or if this mutation was prevalent in H. pylori strains that result in gastric cancer simply by chance.
To investigate this, the authors first looked at the proteinase activity between the gastric cancer strains and the strains from non-cancer patients. When they compared the two, they found that strains with the 171L HtrA mutation (amino acid 171 as a leucine) had more proteinase activity compared to strains without the HtrA mutation (amino acid 171 as a serine). Furthermore, they found that the H. pylori gastric cancer strains degraded the tight junctions of gastric cells more than the non-cancer H. pylori strains. This was significant evidence for the authors that the 171L HtrA mutation observed in clinical isolates of H. pylori from gastric cancer patients was promoting tight junction degradation and overall cancer progression.
Next, the authors wanted to see if the 171L variant of HtrA was able to degrade tight junction proteins specifically. To do this, they used fluorescent microscopy to visualize the tight junctions in 171L variants of HtrA versus 171S variants of HtrA. Indeed, they saw that the levels of occludin, a specific protein that maintains the strength of tight junctions, were decreased in the 171L HtrA variants compared to the 171S variants. This gave the authors great evidence to conclude that the HtrA 171L variant degraded tight junctions in stomach cells (Figure 1).
Correlating the bacteria mutation to cancer:
Now that the authors knew that the 171L variant of HtrA had higher protease activity, they wanted to determine if this variant resulted in other cancer-related phenotypes. First, they examined the ability of the clinical isolates of H. pylori to induce chronic inflammation. Strains with the 171L variant of HtrA had higher levels of various immune response activators, particularly the pro-inflammatory cytokine interleukin-8.
Furthermore, the authors also examined the ability of these strains to induce DNA breaks in the host cells. DNA damage is frequently associated with cancer, as double-stranded DNA breaks can often lead to even more mutations as host proteins that fix the breaks, do not always perfectly fix the sequence of the DNA. As they predicted, the strains of H. pylori with the 171L HtrA variant caused significant host DNA damage (Figure 2). These experiments gave the authors even more evidence that the 171L variant of HtrA promoted cancer.
Where do we go from here?
The findings of this paper directly correlated a single mutation in HtrA at amino acid 171 with gastric cancer development. Strains with this mutation greatly affected the function of this protein. Importantly for cancer progression, this mutation had more degradation of tight junctions and chronic inflammation compared to strains that did not have this mutation. Furthermore, H. pylori strains with this mutation caused an increase in double stranded DNA breaks, which is a key indicator of cancer development. Overall, more studies are needed to determine if having a strain with this mutation long-term is more likely to result in cancer. However these studies provide a solid stepping stone to the long journey of cancer research ahead.
Featured image: Made by author with BioRender.