Breaking down the microbiology world one bite at a time
AI meets virology: Are new drugs just one click away?
The first two weeks of October are exciting and nerve-wracking times for scientists across the globe: The awardees of the prestigious Nobel Prize are announced. This year’s Nobel Prize in Chemistry constitutes a novelty, albeit likely not an exception in the foreseeable future. For the first time, researchers were awarded for a scientific breakthrough achieved by the use of artificial intelligence (AI). Next to David Baker and his work on bottom-up protein design, John Jumper and Demis Hassabis from Google DeepMind in London were recognized for their achievements in AI-assisted protein structure prediction.
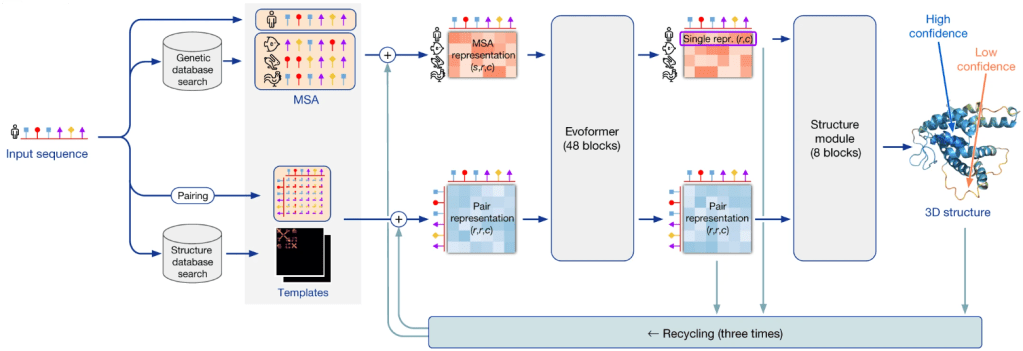
Since its launch in 2020, the AlphaFold algorithm has impacted the work of virtually every biological scientist by predicting three-dimensional protein structures with unmatched accuracy.
Proteins are essential building blocks of life and engage in a myriad of functions inside and outside of our bodies.They are built from 20 amino acids, which are connected in a line (a polypeptide), just like a string of pearls. But it is not as simple as that: Individual pearls (the amino acids), located far apart on the string, interact with one another. Consequently, the string folds into a three-dimensional shape, unique to every protein.
Until the launch of AlphaFold, scientists had to use tedious and time-consuming methods such as X-ray crystallography, cryo-electron microscopy, or nuclear magnetic resonance spectroscopy to determine three-dimensional protein structures. They provide useful information about a protein’s function and ways to target the protein in case of disease. What researchers needed months and years for, AlphaFold can accomplish in a few minutes, feeding on all previously acquired information.
A recent study conducted by researchers from the University of Glasgow applied AI-assisted protein structure prediction to a virological question for the first time. The researchers gained new insights into the evolutionary history of an entire virus family and identified a novel entry mechanism which may aid the development of antivirals and vaccines in the future.
Viruses are generally classified into families based on morphology, composition, and mode of multiplication. The diverse Flaviviridae virus family comprises a large number of viruses besides the well-known hepatitis C virus. The relationships within the same family are usually established by comparing the genomic sequence of a single protein that is relatively similar across the viruses. However, this only offers a very restricted view on virus evolution and does not allow comparison of less conserved viral proteins.
Glycoproteins are the docking points on the surface of a virus and make contact with a host cell. They determine the animals and specific tissues infected by the virus and the mechanism by which the virus enters its host cell. However, such glycoproteins are not well conserved and thus, poorly characterized for many viruses in the Flaviviridae family.
The authors of the study compared the glycoprotein sequences of more than 400 different viruses of the Flaviviridae family with AI-assisted protein structure prediction. This endeavor would be virtually impossible by traditional methods or comparison of genomic sequences. The authors found similarities between glycoproteins of viral subgroups which had not been closely associated with each other previously. They even suggested novel functional mechanisms for the glycoproteins of those subgroups, including hepatitis C virus. This changes our understanding of how these viruses make contact with cells, which can help the identification of therapeutic targets in the future. The authors also redefined the evolutionary history of the entire Flaviviridae family based on their AI-predicted structural similarities.
This study demonstrates how the power of AI tools can massively boost scientific progress and allow researchers to advance to more pressing questions concerning, for example, disease control and pandemic preparedness. It is becoming clearer and clearer: The future of research is unimaginable without AI-informed tools.
Link to the original post: Mifsud, J.C.O., Lytras, S., Oliver, M.R. et al. Mapping glycoprotein structure reveals Flaviviridae evolutionary history. Nature 633, 695–703 (2024). https://doi.org/10.1038/s41586-024-07899-8
Featured image: Image source: Created using Bing Image Creator
Additional sources: https://deepmind.google/discover/blog/alphafold-a-solution-to-a-50-year-old-grand-challenge-in-biology/