Breaking down the microbiology world one bite at a time
Detection of CRKP infections using CRISPR-Cas9
A gram-negative pathogenic bacterium called Klebsiella Pneumoniae (KP) is one of the leading causes of hospital-acquired infections and community-acquired infections (CA), such as bloodstream infection, pneumonia and urinary tract infection. In hospitals, one of the main sources of transmission for KP is person-to-person contact between patients and healthcare workers; with hands by healthcare workers being the main form of transmissions.
The most common infection site for KP is the urinary tract due environmental conditions that allows the bacterium to freely colonise. KP is frequently found in water, soil, sewage and plant surfaces. Hence, once a person has transmitted the KP, the bacterium grows in humans on surfaces called the mucosal surface; an example is the nasopharynx – which is the top area of the throat. However, the main issue with KP is that the bacterium is increasingly resistant towards antibiotics, as well as containing a mechanism that has made the bacterium multi-drug resistant towards various antimicrobial classes. Common ß-Lactam drugs such as Carbapenem antibiotics, which are considered as “last line agents” for medical treatments dealing with severe infections caused by gram-negative bacterium, are gradually becoming less effective due to carbapenem-resistant KP (CRKP) strains. Thus, causing a global public health issue. Therefore, are CRKP strains currently undefeated by antibiotics or is there another form of treatment that can conquer the bacterium?
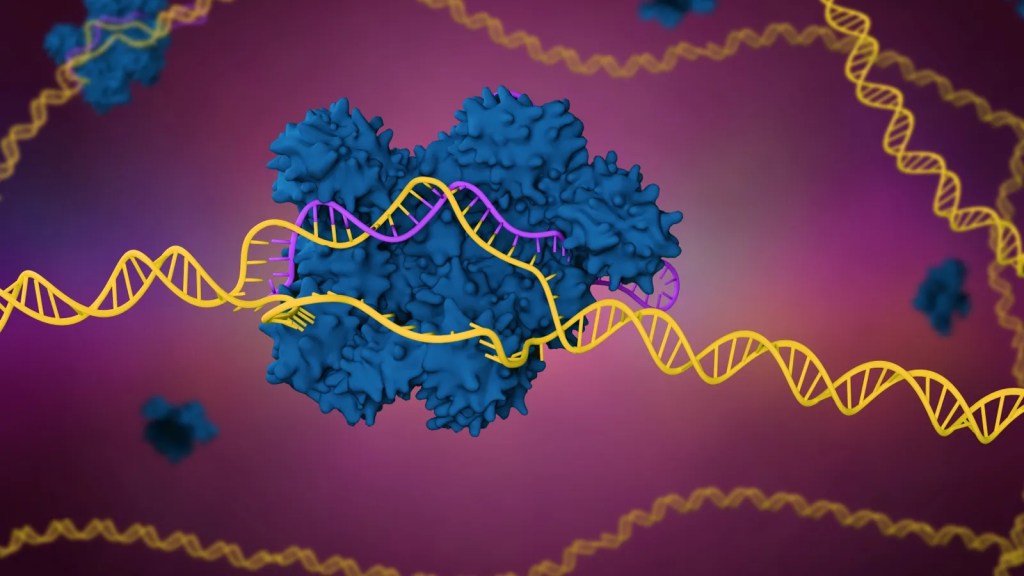
Over the years, scientists have been able to use a unique form of technology to remove, edit and alter sections within DNA; this includes using the technology towards micro-organisms such as bacteria and viruses. CRISPR is an example of genome editing technology which consists of a protein that is used to modify or mutate DNA. Cas9 proteins are widely used by scientists, especially since the protein can be programmed target and bind to specific DNA sequences; this is done through the guidance of a single strand DNA, also known as RNA The Cas9 protein attaches to the RNA, which then moves along the DNA strand until it detects and binds to a 20-base-pair-sequence, also known as a 20-nt guide sequence, that matches the RNA sequence. Once the sequence has been detected the Cas9 protein cuts the targeted DNA sequence, and the gene is repaired or modified.
So, if CRISPR-Cas method is used for genome editing, the question to be asked is how effective is the technology at editing or modifying microbial genes like KP?
According to a study conducted in 2023, CRISPR-Cas13a combined with another detection system called PCR amplification was used to identify environmental samples of KP using culture medium to obtain single colonies. The outcome of the experiment revealed that after 24 hours the CRISPR-Cas13a system was able to detect a low concentration of samples with a culture colony less than 1 CFU/cm2, even though no colonies were obtained. However, more colonies grew on the medium after 48 hours.
| Genome Editing Technology | Number of detected CRKP Strains | Probability to detect CRKP strains |
| PCR-CRISPR-Cas13a assay | Positive: 47 | 92.16% |
| Negative: 4 | ||
| RAA-CRISPR-Cas13a assay | Positive: 47 | 92.16% |
| Negative: 4 |
The study also compared the effectiveness of detection by PCR-CRISPR-Cas13a with another amplification CRISPR system (recombinase-aided amplification or RAA). For this comparison, 61 clinical strains were collected and 51 of those strains were identified as CRKP. From Table 1, the results show that PCR and the RAA detected 47 CRKP gram-positive strains and 4 gram-negative strains. PCR also showed a 92.16% probability of detecting positive samples, also known as positive coincidence rate. Therefore, the study proved that the CRISPR-Cas13a system can be used to improve the monitory process of antimicrobial resistance. Even though the genome technique could show some challenges for critically ill patients to obtain effective antibiotics within the first 24 hours due to the need for growth of approximately 48 hours in culture medium. However, the CRISPR-Cas13a system can also assist patients with acquiring the correct prescriptions and treatment to avoid the KP infections from spreading any further.
CRISPR shows potential and a new path for assisting medical monitoring due to the system’s high sensitivity for detection. Despite CRISPR’s flaws for processing a 24-hour diagnosis, this technique has shown to be effective with further research and improvement.
Featured image: BioPharma Dive. (2024). Gene editing biotechs face new uncertainty after CRISPR patent ruling. [online] Available at: https://www.biopharmadive.com/news/crispr-patent-biotech-uc-broad-intellia/619684/.