Breaking down the microbiology world one bite at a time
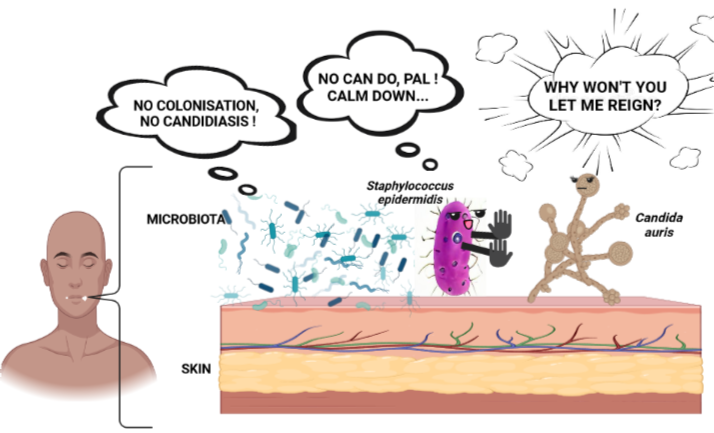
The skin’s “frenemy”
Research advancements on human microbiota, comprising the trillions of microbes that live in and on us, have changed how scientists and clinicians understand the development and progression of many human diseases. The skin is no exception. Scientists are now learning about the microbial components of healthy skin, how these microbes provide essential functions to our skin cells and immune system, and importantly, how their partnerships are disrupted and turn hostile in various skin diseases.
Atopic dermatitis, commonly known as eczema, is a ubiquitous inflammatory skin disease (1). Think: chronically inflamed, red, itchy, dry skin. The root causes of eczema are unclear, and likely a consequence of the complex interplay between our genetics, environment, immune function, and skin microbes. Regarding the latter, the skin microbe Staphylococcus aureus sits at the center of attention for eczema pathologists (2). However, recent research has revealed that another possible culprit in this disease’s severity is, surprisingly, the skin commensal Staphylococcus epidermidis (3). S. epidermidis is commonly regarded as a crucial member of healthy skin—and it is. However, S. epidermidis is, perhaps, like the microbial Jekyll and Hyde: In some instances, the symbiont is protective against severe skin infection, and in general, benefits skin homeostasis and integrity. By contrast, in other instances, it can exacerbate skin barrier disruption (4). A persistent challenge in the field is pinpointing the environments that favor S. epidermidis commensalism versus S. epidermidis pathogenicity.
In a recent study, researchers honed in on the hypothesis that, in eczema patients, S. epidermidis is making molecules that damage the skin (3). Specifically, they examined the role of proteases, extracellular enzymes that chew up proteins. They identified a protease called EcpA, which they discovered could degrade an essential skin barrier protein and skin-produced anti-microbial. To test the capacity of EcpA to disrupt the epidermal barrier, researchers colonized mouse skin with two distinct strains of S. epidermidis: one that makes EcpA, and one that does not. Only the S. epidermidis strain that produced EcpA could penetrate the skin barrier and drive skin damage. Indeed, application of purified EcpA protein directly to mouse skin resulted in a similar skin barrier disruption, implicating a direct role for EcpA in exacerbating barrier degradation.

Interestingly, the gene that codes for EcpA is present in commensal and pathogenic S. epidermidis strains, suggesting that there could be specific conditions that enable EcpA-mediated virulence. First, the researchers observe that not all S. epidermidis produce EcpA to the same level. Thus, strain-level diversity, particularly, in terms of EcpA expression, could be a key signature to predicting disease progression. Second, EcpA expression is regulated by a bacterial signaling process called quorum sensing. Quorum sensing is a type of cell-cell communication that allows bacteria to take a census of the density and composition of their vicinal community and coordinate their gene expression accordingly. In this case, at high S. epidermidis cell density, EcpA production accelerates. The ramification of this is that EcpA is maximally expressed in skin microbial communities that harbor an S. epidermidis overgrowth. Taken together, the results of this study indicate that skin damage in eczema patients could be intensified by S. epidermidis strains that naturally make more EcpA, and imbalanced community conditions (i.e. high S. epidermidis abundance) that favor the expression of this deleterious protein.

Our skin microbiome operates on a delicate system of checks and balances. Tipping the scales in one direction (i.e., too much of one bacterial species, too little of another, a decline in species diversity) could wreak havoc on normal skin functions and stoke immune responses. Indeed, microbial imbalance, or dysbiosis, is a common thread among many inflammatory diseases. As noted, the drivers of eczema are multifaceted and complex. Now that scientists are figuring out how S. epidermidis could be virulent in cases of eczema, the next step is to identify how additional factors (perhaps, genetic or environmental) coalesce to facilitate the ideal environment for S. epidermidis pathogenicity.
While the onset and severity of eczema cannot solely be attributed to S. epidermidis, pinpointing specific agents of skin damage, like EcpA, could inform more highly-selective, personalized approaches to treatment and symptom management that do not interfere with host immune function. For example, innovative medicines could antagonize S. epidermidis quorum sensing to inhibit production of EcpA. Anti-quorum sensing molecules have already been shown to diminish S. aureus-driven inflammation and skin damage (5). By contrast, the hallmark of existing eczema treatments are broad-spectrum corticosteroids and other immunosuppressants (6), which dampen the host immune response. The drawbacks of such drugs are that they do not treat microbial dysbiosis, and they could confer serious side-effects to our overall physiology in the long-term (7). Perhaps, focusing on the microbial components of skin diseases could become a mainstay in future development of safe and effective therapeutics. More broadly, knowledge of how microbial homeostasis is established (and disrupted) could launch novel approaches to standard care and maintenance of healthy skin.
References:
6. Plant A, Ardern-Jones MR. Advances in atopic dermatitis. Clin Med. 2021 May 17;21(3):177–81.
Featured Image: https://www.wannapik.com/vectors/28885